Encoding binary data into DNA sequence
post, Jan 3, 2019, on Mitja Felicijan's blog
Initial thoughts
Imagine a world where you could go outside and take a leaf from a tree and put
it through your personal DNA sequencer and get data like music, videos or
computer programs from it. Well, this is all possible now. It was not done on a
large scale because it is quite expensive to create DNA strands but it's
possible.
Encoding data into DNA sequence is relatively simple process once you understand
the relationship between binary data and nucleotides and scientists have been
making large leaps in this field in order to provide viable long-term storage
solution for our data that would potentially survive our specie if case of
global disaster. We could imprint all the world's knowledge into plants and
ensure the survival of our knowledge.
More optimistic usage for this technology would be easier storage of ever
growing data we produce every day. Once machines for sequencing DNA become fast
enough and cheaper this could mean the next evolution of storing data and
abandoning classical hard and solid state drives in data warehouses.
As we currently stand this is still not viable but it is quite an amazing and
cool technology.
My interests in this field are purely in encoding processes and experimental
testing mainly because I don't have the access to this expensive machines. My
initial goal was to create a toolkit that can be used by everybody to encode
their data into a proper DNA sequence.
Glossary
deoxyribose A five-carbon sugar molecule with a hydrogen atom rather than a
hydroxyl group in the 2′ position; the sugar component of DNA nucleotides.
double helix The molecular shape of DNA in which two strands of nucleotides
wind around each other in a spiral shape.
nitrogenous base A nitrogen-containing molecule that acts as a base; often
referring to one of the purine or pyrimidine components of nucleic acids.
phosphate group A molecular group consisting of a central phosphorus atom
bound to four oxygen atoms.
RGB The RGB color model is an additive color model in which red, green and
blue light are added together in various ways to reproduce a broad array of
colors.
GCC The GNU Compiler Collection is a compiler system produced by the GNU
Project supporting various programming languages.
Data encoding
TL;DR: Encoding involves the use of a code to change original data into a
form that can be used by an external process.
Encoding is the process of converting data into a format required for a number
of information processing needs, including:
- Program compiling and execution
- Data transmission, storage and compression/decompression
- Application data processing, such as file conversion
Encoding can have two meanings:
- In computer technology, encoding is the process of applying a specific code,
such as letters, symbols and numbers, to data for conversion into an
equivalent cipher.
- In electronics, encoding refers to analog to digital conversion.
Quick history of DNA
- 1869 - Friedrich Miescher identifies "nuclein".
- 1900s - The Eugenics Movement.
- 1900 – Mendel's theories are rediscovered by researchers.
- 1944 - Oswald Avery identifies DNA as the 'transforming principle'.
- 1952 - Rosalind Franklin photographs crystallized DNA fibres.
- 1953 - James Watson and Francis Crick discover the double helix structure of DNA.
- 1965 - Marshall Nirenberg is the first person to sequence the bases in each codon.
- 1983 - Huntington's disease is the first mapped genetic disease.
- 1990 - The Human Genome Project begins.
- 1995 - Haemophilus Influenzae is the first bacterium genome sequenced.
- 1996 - Dolly the sheep is cloned.
- 1999 - First human chromosome is decoded.
- 2000 – Genetic code of the fruit fly is decoded.
- 2002 – Mouse is the first mammal to have its genome decoded.
- 2003 – The Human Genome Project is completed.
- 2013 – DNA Worldwide and Eurofins Forensic discover identical twins have differences in their genetic makeup.
What is DNA?
Deoxyribonucleic acid, a self-replicating material which is present in nearly
all living organisms as the main constituent of chromosomes. It is the
carrier of genetic information.
The nitrogen in our DNA, the calcium in our teeth, the iron in our blood,
the carbon in our apple pies were made in the interiors of collapsing stars.
We are made of starstuff.
-- Carl Sagan, Cosmos
The nucleotide in DNA consists of a sugar (deoxyribose), one of four bases
(cytosine (C), thymine (T), adenine (A), guanine (G)), and a phosphate.
Cytosine and thymine are pyrimidine bases, while adenine and guanine are purine
bases. The sugar and the base together are called a nucleoside.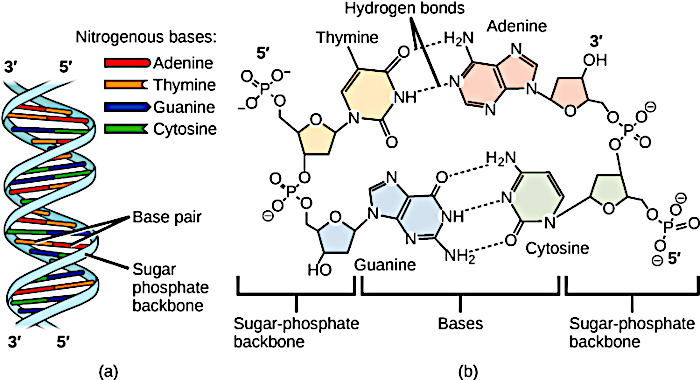
DNA (a) forms a double stranded helix, and (b) adenine pairs with thymine and
cytosine pairs with guanine. (credit a: modification of work by Jerome Walker,
Dennis Myts)
Encode binary data into DNA sequence
As an input file you can use any file you want:
- ASCII files,
- Compiled programs,
- Multimedia files (MP3, MP4, MVK, etc),
- Images,
- Database files,
- etc.
Note: If you would copy all the bytes from RAM to file or pipe data to file you
could encode also this data as long as you provide file pointer to the encoder.
Basic Encoding
As already mentioned, the Basic Encoding is based on a simple mapping. Since DNA
is composed of 4 nucleotides (Adenine, Cytosine, Guanine, Thymine; usually
referred using the first letter). Using this technique we can encode
using a single nucleotide. In this way, we are able to use the 4 bases that
compose the DNA strand to encode each byte of data.
| Two bits | Nucleotides |
|---|
| 00 | A (Adenine) |
| 10 | G (Guanine) |
| 01 | C (Cytosine) |
| 11 | T (Thymine) |
With this in mind we can simply encode any data by using two-bit to Nucleotides
conversion.
{ Algorithm 1: Naive byte array to DNA encode }
procedure EncodeToDNASequence(f) string
begin
enc string
while not eof(f) do
c byte := buffer[0] { Read 1 byte from buffer }
bin integer := sprintf('08b', c) { Convert to string binary }
for e in range[0, 2, 4, 6] do
if e[0] == 48 and e[1] == 48 then { 0x00 - A (Adenine) }
enc += 'A'
else if e[0] == 48 and e[1] == 49 then { 0x01 - G (Guanine) }
enc += 'G'
else if e[0] == 49 and e[1] == 48 then { 0x10 - C (Cytosine) }
enc += 'C'
else if e[0] == 49 and e[1] == 49 then { 0x11 - T (Thymine) }
enc += 'T'
return enc { Return DNA sequence }
end
Another encoding would be Goldman encoding. Using this encoding helps with
Nonsense mutation (amino acids replaced by a stop codon) that occurs and is the
most problematic during translation because it leads to truncated amino acid
sequences, which in turn results in truncated proteins.
Where to store big data? In DNA: Nick Goldman at TEDxPrague
In bioinformatics, FASTA format is a text-based format for representing either
nucleotide sequences or peptide sequences, in which nucleotides or amino acids
are represented using single-letter codes. The format also allows for sequence
names and comments to precede the sequences. The format originates from the
FASTA software package, but has now become a standard in the field of
bioinformatics.
The first line in a FASTA file started either with a ">" (greater-than) symbol
or, less frequently, a ";" (semicolon) was taken as a comment. Subsequent lines
starting with a semicolon would be ignored by software. Since the only comment
used was the first, it quickly became used to hold a summary description of the
sequence, often starting with a unique library accession number, and with time
it has become commonplace to always use ">" for the first line and to not use
";" comments (which would otherwise be ignored).
;LCBO - Prolactin precursor - Bovine
; a sample sequence in FASTA format
MDSKGSSQKGSRLLLLLVVSNLLLCQGVVSTPVCPNGPGNCQVSLRDLFDRAVMVSHYIHDLSS
EMFNEFDKRYAQGKGFITMALNSCHTSSLPTPEDKEQAQQTHHEVLMSLILGLLRSWNDPLYHL
VTEVRGMKGAPDAILSRAIEIEEENKRLLEGMEMIFGQVIPGAKETEPYPVWSGLPSLQTKDED
ARYSAFYNLLHCLRRDSSKIDTYLKLLNCRIIYNNNC*
>MCHU - Calmodulin - Human, rabbit, bovine, rat, and chicken
ADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTID
FPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREA
DIDGDGQVNYEEFVQMMTAK*
>gi|5524211|gb|AAD44166.1| cytochrome b [Elephas maximus maximus]
LCLYTHIGRNIYYGSYLYSETWNTGIMLLLITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTNLV
EWIWGGFSVDKATLNRFFAFHFILPFTMVALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLG
LLILILLLLLLALLSPDMLGDPDNHMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALFLSIVIL
GLMPFLHTSKHRSMMLRPLSQALFWTLTMDLLTLTWIGSQPVEYPYTIIGQMASILYFSIILAFLPIAGX
IENY
FASTA format was extended by FASTQ
format from the Sanger Centre in Cambridge.
PNG encoded DNA sequence
| Nucleotides | RGB | Color name |
|---|
| A ➞ Adenine | (0,0,255) | Blue |
| G ➞ Guanine | (0,100,0) | Green |
| C ➞ Cytosine | (255,0,0) | Red |
| T ➞ Thymine | (255,255,0) | Yellow |
With this in mind we can create a simple algorithm to create PNG representation
of a DNA sequence.
{ Algorithm 2: Naive DNA to PNG encode from FASTA file }
procedure EncodeDNASequenceToPNG(f)
begin
i image
while not eof(f) do
c char := buffer[0] { Read 1 char from buffer }
case c of
'A': color := RGB(0, 0, 255) { Blue }
'G': color := RGB(0, 100, 0) { Green }
'C': color := RGB(255, 0, 0) { Red }
'T': color := RGB(255, 255, 0) { Yellow }
drawRect(i, [x, y], color)
save(i) { Save PNG image }
end
Encoding text file in practice
In this example we will take a simple text file as our input stream for
encoding. This file will have a quote from Niels Bohr and saved as txt file.
How wonderful that we have met with a paradox. Now we have some hope of
making progress.
― Niels Bohr
First we encode text file into FASTA file.
./dnae-encode -i quote.txt -o quote.fa
2019/01/10 00:38:29 Gathering input file stats
2019/01/10 00:38:29 Starting encoding ...
106 B / 106 B [==================================] 100.00% 0s
2019/01/10 00:38:29 Saving to FASTA file ...
2019/01/10 00:38:29 Output FASTA file length is 438 B
2019/01/10 00:38:29 Process took 987.263µs
2019/01/10 00:38:29 Done ...
Output of quote.fa file contains the encoded DNA sequence in ASCII format.
>SEQ1
GACAGCTTGTGTACAAGTGTGCTTGCTCGCGAGCGGGTACGCGCGTGGGCTAACAAGTGA
GCCAGCAGGTGAACAAGTGTGCGGACAAGCCAGCAGGTGCGCGGACAAGCTGGCGGGTGA
ACAAGTGTGCCGGTGAGCCAACAAGCAGACAAGTAAGCAGGTACGCAGGCGAGCTTGTCA
ACTCACAAGATCGCTTGTGTACAAGTGTGCGGACAAGCCAGCAGGTGCGCGGACAAGTAT
GCTTGCTGGCGGACAAGCCAGCTTGTAAGCGGACAAGCTTGCGCACAAGCTGGCAGGCCT
GCCGGCTCGCGTACAAATTCACAAGTAAGTACGCTTGCGTGTACGCGGGTATGTATACTC
AACCTCACCAAACGGGACAAGATCGCCGGCGGGCTAGTATACAAGAACGCTTGCCAGTAC
AACC
Then we encode FASTA file from previous operation to encode this data into PNG.
./dnae-png -i quote.fa -o quote.png
2019/01/10 00:40:09 Gathering input file stats ...
2019/01/10 00:40:09 Deconstructing FASTA file ...
2019/01/10 00:40:09 Compositing image file ...
424 / 424 [==================================] 100.00% 0s
2019/01/10 00:40:09 Saving output file ...
2019/01/10 00:40:09 Output image file length is 1.1 kB
2019/01/10 00:40:09 Process took 19.036117ms
2019/01/10 00:40:09 Done ...
After encoding into PNG format this file looks like this.
The larger the input stream is the larger the PNG file would be.
Compiled basic Hello World C program with
GCC would look
like.
// gcc -O3 -o sample sample.c
#include <stdio.h>
main() {
printf("Hello, world!\n");
return 0;
}
I have created a toolkit with two main programs:
- dnae-encode (encodes file into FASTA file)
- dnae-png (encodes FASTA file into PNG)
Toolkit with full source code is available on
github.com/mitjafelicijan/dna-encoding.
dnae-encode
> ./dnae-encode --help
usage: dnae-encode --input=INPUT [<flags>]
A command-line application that encodes file into DNA sequence.
Flags:
--help Show context-sensitive help (also try --help-long and --help-man).
-i, --input=INPUT Input file (ASCII or binary) which will be encoded into DNA sequence.
-o, --output="out.fa" Output file which stores DNA sequence in FASTA format.
-s, --sequence=SEQ1 The description line (defline) or header/identifier line, gives a name and/or a unique identifier for the sequence.
-c, --columns=60 Row characters length (no more than 120 characters). Devices preallocate fixed line sizes in software.
--version Show application version.
dnae-png
> ./dnae-png --help
usage: dnae-png --input=INPUT [<flags>]
A command-line application that encodes FASTA file into PNG image.
Flags:
--help Show context-sensitive help (also try --help-long and --help-man).
-i, --input=INPUT Input FASTA file which will be encoded into PNG image.
-o, --output="out.png" Output file in PNG format that represents DNA sequence in graphical way.
-s, --size=10 Size of pairings of DNA bases on image in pixels (lower resolution lower file size).
--version Show application version.
Benchmarks
First we generate some binary sample data with dd.
dd if=<(openssl enc -aes-256-ctr -pass pass:"$(dd if=/dev/urandom bs=128 count=1 2>/dev/null | base64)" -nosalt < /dev/zero) of=1KB.bin bs=1KB count=1 iflag=fullblock

Our freshly generated 1KB file looks something like this (its full of
garbage data as intended).
We create following binary files:
- 1KB.bin
- 10KB.bin
- 100KB.bin
- 1MB.bin
- 10MB.bin
- 100MB.bin
After this we create FASTA files for all the binary files by encoding them
into DNA sequence.
./dnae-encode -i 100MB.bin -o 100MB.fa
Then we GZIP all the FASTA files to see how much the can be compressed.
gzip -9 < 10MB.fa > 10MB.fa.gz

The speed increase that occurs when encoding to FASTA format.
Size of the out file after encoding.
Download CSV file with benchmarks.
References